Our Research
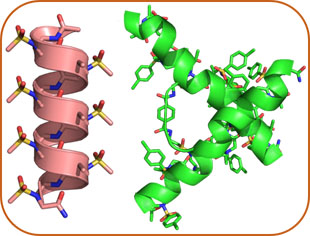
AApeptide Folding
We recently developed a new class of sequence-specific peptidomimetics - AApeptides - inspired by chiral PNA backbones. AApeptides fold into well-defined protein-like secondary and tertiary structures. Building on these results, we aim to design hierarchical AApeptide structures with predictable functions, such as artificial proteins with enzymatic activity.
Examples:
Angew. Chem. Int. Ed., 2019;
Angew. Chem. Int. Ed., 2018;
J. Am. Chem. Soc., 2018;
J. Am. Chem. Soc., 2017
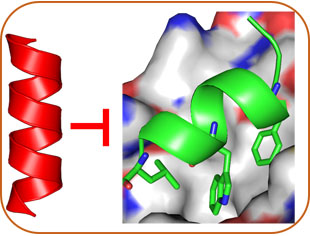
Structure-Based Design
Our lab develops molecular probes and drug candidates using Rational Design and Combinatorial Screening. With stable and diverse helical AApeptides, we design mimetics to modulation protein interactions to combat cancer, Alzheimer's disease, Parkinson's disease, diabetes, SARS-CoV-2, HIV and bacterial infections.
Examples:
J. Am. Chem. Soc., 2025;
J. Med. Chem., 2025;
PNAS, 2024;
J. Am. Chem. Soc., 2023;
ACS Cent. Sci., 2023;
J. Am. Chem. Soc., 2022;
Sci. Adv., 2020;
PNAS, 2019
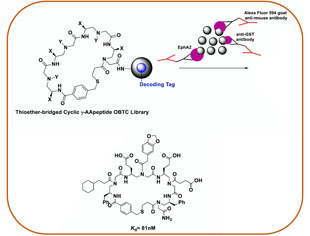
Combinatorial Screening
As a complement to Structure-Based Design, we also employ combinatorial library screening. By creating diverse AApeptide libraries, we identify selective binders for proteins and biomacromolecules that can modulate biological functions. These hits may serve as promising drug leads or candidates.
Examples:
Cell Dis., 2022;
Acta Pharm. Sin. B, 2021;
J. Med. Chem., 2021;
Cell Chem. Biol., 2021;
J. Med. Chem., 2017
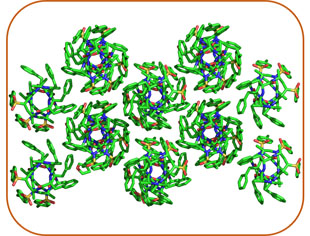
Supramolecular Chemistry
AApeptides can form defined structures that assemble into functional supramolecular materials when tailored with specific side chains. We design and synthesize diverse AApeptide-based assemblies to explore their potential in biomaterials and nanotechnology applications.
Examples:
Nat. Commun., 2025;
J. Am. Chem. Soc., 2019
